Gene Sequencing involves detecting the DNA building blocks (nucleotides) of an individual’s genetic code. Sanger sequencing was originally used but is time consuming and expensive and it would take years to sequence all of a person’s DNA- known as their Genome. Next-Generation sequencing has sped up the process, taking only days to weeks to sequence a whole genome.
Single Gene Analysis
This is genotyping for a specific chosen gene only: originally mainly done before Panel tests were developed. It is still used for testing family members of someone (the proband), who has been found to have a gene variant.
Panel Testing
Panel testing refers to the testing of groups of genes usually associated with particular medical conditions. In The London Genetics Centre Study we are doing two large panel tests, cancer and cardiac.
Cancer panels looks for germline cancer predispositions that are known to be associated with a particular cancer. They have the special gain of not missing large deletions, which can very rarely happen with WGS, but they do not have the massive wide gene coverage of WGS.
The landscape of genetic testing has shifted from single gene testing to that of multi-gene panel opened up by the continued decline in cost. In many cancer cases gene panels testing for germline genetic alterations that can cause that specific cancer are ordered. For instance, a breast, ovarian or colorectal panel of genes. Alternatively, a cardiac gene panel.
The latest Breast Panels now include: BRCA 1/2, PALB2, ATM, CHEK2, PTEN, STK11, TP53 and BARD1. (The NHS currently only tests for BRCA and PALB2 alterations).
In the UK, the National Institute of Health and Care excellence, sets a cost effectiveness threshold of gene testing for family history situations, where there is a pre-test 10% or greater estimated likelihood of finding a significant genetic alteration.
Whole Exome Screening
Sequencing for the DNA that provides instructions for making proteins. These pieces called exons are thought to make up 1% of a person’s genome. It is an efficient method for identifying disease causing mutations. Analysing coding areas of all genes, it will include genes such as cystic fibrosis and haemochromatosis as with WGS. It will not enable you do the SNPs nor calculate polygenic risk scores which is possible with WGS. It could also miss some carrier genes, if they are in the intronic regions.
Whole Genome Sequencing
DNA Sequencing for all the nucleotides and their order.
This will also detect DNA variations outside the Exons which are now known to affect gene activity and protein production leading to genetic disorders.
At The London Genetics Centre we are offering Whole Genome Screening. We obtain all the genetic code, so with your permission, we can then send it to Prof Eeles’ Institute of Cancer Research’s Laboratory at Sutton to do single nucleotide polymorphism (SNP) profiling. SNP profiling can be done for different conditions such as prostate cancer, (where Prof Eeles has discovered 2/3rds of the worlds SNPs). We are also doing SNP profiling for Breast, Ovarian and Colorectal cancers. Cardiovascular SNP risk profiling is also being carried out to detect those at higher risk of cardiovascular disease, some of whom are not identifiable by the usual cardiovascular risk factors. At present this is in the research arena but we hope that in the near future this will be important for personal healthcare risk identification with consequent preventative actions.
Variant Classification
The American College of Medical Genetics and Genomics (ACMG) grades variants into 5 categories. Class 5 Pathogenic – disease causing, Class 4 -likely pathogenic >90% certainty of the variant being disease causing. Class 3 is a variant of uncertain significance (VUS). Typically most VUS’s have about a one in a hundred chance of ever being upgraded with time to pathogenic; so some organisations do not report them as in the vast majority they cause unnecessary costs and potentially worry.
Whole Genome Sequencing leading to Polygenic Risk Scores with SNP profiling detail.
With Whole Genome Screening by adding all the SNP scores for a particular condition we can produce polygenic risk scores for specific conditions identifying those with the highest scores. For example, a person with a polygenic risk score for prostate cancer in the top 5% of the risk profile will have 6 times the risk of prostate cancer compared with the average of the population. Such individuals can be offered research studies investigating the role of targeted screening in such groups.
In summary, considering prostate cancer the genetic screening system at The London Genetics Centre will look for 8 major genes for prostate cancer of which BRCA genes are one part. We will also do Prostate SNP profiling to give a prostate polygenic risk score, picking up further high-risk men, extending beyond those identified by the major genes.
An example for prostate cancer using WGS data to provide a Polygenic risk score (PRS)
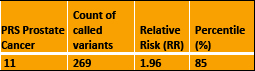
PRS Prostate Cancer Count of called variants Relative Risk (RR) Percentile (%)
• The Result means the risk of prostate cancer is about 2-fold higher (RR=1.96) compared to the general UK /European ancestry population risk.
• If we were to use PRS in the future, as a prostate cancer screening tool it may be that screening men in the top 10% of the risk distribution will be advised.
• This result at 85th centile does not fall into this high-risk category.
• However, if future studies advise screening the top 20% of the population, this person would then fall into the higher-risk category.
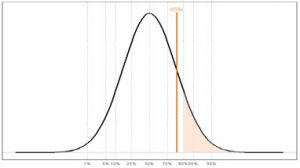
Prostate Genetic Risk Distribution
Pharmacogenes
With our whole genome screening we are also able to do the Pharmacogenes showing how we individually metabolise drugs. This is a new and developing area of medicine. Medicines and their dosing can be personally tailored for you. Also, drugs that could be potentially fatal for you, or on the other hand not effective can be avoided